The primary goal of the Human Genome Project was to provide a comprehensive and accurate reference map of the human genome. This would enable researchers to better understand the genetic basis of human biology, development, and disease. By decoding the complete sequence of DNA, scientists aimed to identify and catalog all the genes in the genome, determine their function, and investigate their role in health and disease.
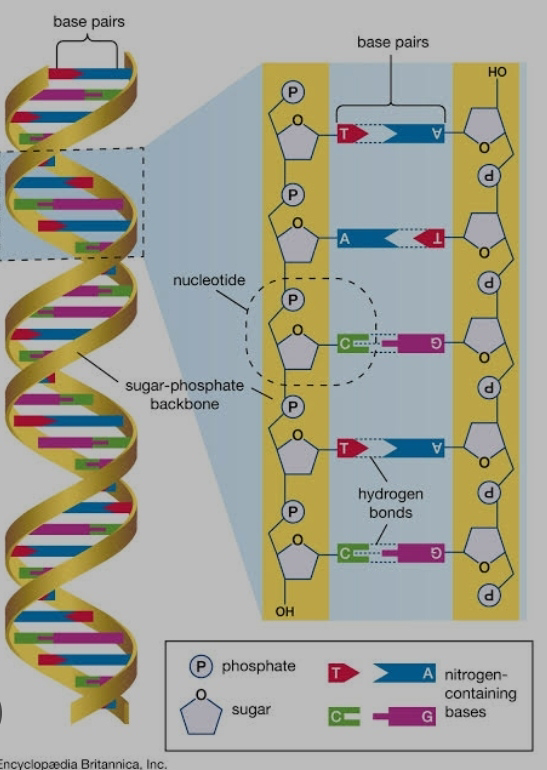
The human genome is composed of about 3 billion base pairs, which are the building blocks of DNA. These base pairs are arranged in a unique sequence that contains both coding and non-coding regions. The coding regions, known as genes, provide instructions for producing proteins, the molecules that carry out most of the biochemical functions in the body. Non-coding regions, although not directly involved in protein production, play important regulatory roles in gene expression and other cellular processes.
Sequencing the human genome was an enormous technical challenge. The traditional methods used to sequence DNA were time-consuming and expensive. To overcome these limitations, the Human Genome Project employed a revolutionary approach called the shotgun sequencing method. This method involved breaking the genome into small fragments, sequencing each fragment, and then using advanced computer algorithms to assemble the pieces into a complete sequence.
The sequencing process began with the collection of DNA samples from multiple individuals. These samples were then purified and fragmented into manageable sizes. Next, the fragments were cloned and inserted into bacterial cells, creating libraries of DNA clones. These clones were then sequenced using automated machines that read the sequence of each fragment.
The sequencing data generated by these machines produced short DNA sequences, called reads. The challenge was to align and overlap these reads accurately to reconstruct the original DNA sequence. To achieve this, sophisticated computational algorithms were developed to assemble the reads into longer contiguous sequences, known as contigs. By comparing overlapping contigs, scientists could further assemble them into larger units called scaffolds. Finally, through additional analysis and refinement, the complete genome sequence was obtained.
The availability of the human genome sequence has had a profound impact on various fields of research. One significant outcome is the advancement of personalized medicine. Understanding an individual's genetic makeup allows for the identification of genetic variations associated with disease susceptibility and drug response. This knowledge has paved the way for tailored treatments and therapies, optimizing patient outcomes.
The Human Genome Project also provided invaluable insights into human evolution and the relationships between different species. By comparing the human genome with those of other organisms, scientists have gained a deeper understanding of the common ancestry and evolutionary changes that have shaped life on Earth.
Furthermore, the project has facilitated the discovery of numerous disease-causing genes and mutations. Scientists have been able to identify genes linked to various genetic disorders, such as cystic fibrosis, Huntington's disease, and certain types of cancer. This knowledge has contributed to the development of diagnostic tests, genetic counseling, and potential gene-based therapies.
The impact of the Human Genome Project extends beyond human health. The project has fueled advancements in biotechnology and genomic research. It has provided researchers with an invaluable resource for studying gene function, regulation, and the intricate interplay of genes in complex biological systems. The availability of vast genomic data has also propelled the development of bioinformatics, a field that combines biology and computer science to analyze and interpret large-scale biological data.
In addition to the scientific achievements, the Human Genome Project sparked ethical, legal, and social discussions. The project raised concerns about the privacy and potential misuse of genetic information.







0 Comments